Tutorial¶
This tutorial introduces geomalgo through simple examples, exploring three differents aspects of its API:
- Python API: Manipulate Python objects in the Python interactive interpreter.
- Cython API: Use Cython compiler to add static typing and manipulate C structures.
- Work on large data sets: Use SoA memory layout and compile to C code to efficiently work large date sets.
These concepts will be demonstrated on a simple example: computing intersection between segments.
Using the Python interface¶
This tutorial is written in a Jupyter notebook. pylab is imported, and matplotlib figures are inlined in the notebook. geomalgo is imported as ga.
In [1]:
%pylab inline
import geomalgo as ga
Populating the interactive namespace from numpy and matplotlib
In [2]:
%load_ext Cython
In [3]:
%%cython
cimport geomalgo as ga
cdef:
ga.CSegment2D seg
Creating points and segments¶
The first geomalgo class (actually, extension type) we manipulate is geomalgo.Point2D.
It takes point coordinates as argument. The Python API also accept an optional name, and provides a plot method, usefull to quickly create matplotlib figures.
In [4]:
A = ga.Point2D(2, 1, name='A')
B = ga.Point2D(6, 4, name='B')
C = ga.Point2D(5, 2, name='C')
D = ga.Point2D(2, 3.5, name='D')
for obj in [A, B, C, D]:
obj.plot()
These four points can be used to create two segments, using geomalgo.Segment2D.
Like points, segments have a plot method.
In [5]:
AB = ga.Segment2D(A, B)
CD = ga.Segment2D(C, D)
for obj in [A, B, C, D, AB, CD]:
obj.plot()
Computing intersection¶
The next task we want to achieve is to compute the intersection between the two segments.
There is a Segment2D.intersect_segment() method to compute intersection with another segment. It returns two objects, which can be:
- both None if segments do not intersect,
- a point and None if there is one intersection point,
- two points when segments are colinear.
In [6]:
I0, I1 = AB.intersect_segment(CD)
print(I0)
print(I1)
for obj in [A, B, C, D, AB, CD]:
obj.plot()
I0.plot(name='I', color='red')
Point2D(4.0, 2.5)
None
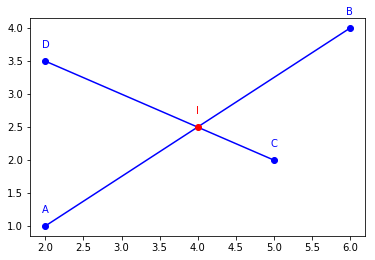
Segment relative coordinates¶
Another usefull information is to locate the intersection point not only with cartesian coordiante,
but also with coordiante relative to each segment. This is achieve with the return_coords flag.
Note that as there is not second intersection point, we can ignore None value, assigning them to a _ variable.
In [7]:
I0, _, (AB_coord, CD_coord, _, _) = AB.intersect_segment(CD, return_coords=True)
print(AB_coord, CD_coord)
0.5 0.3333333333333333
AB_coord is \(\frac{1}{2}\), meaning the point is in the middle of the segment, while CD_coord is \(\frac{1}{3}\), meaning the point is at a distance \(\frac{|CD|}{3}\) from the point C.
To change from cartesian coordinates to segment coordiante, use the Point2D.at() method:
In [8]:
print(AB.at(AB_coord))
print(CD.at(CD_coord))
Point2D(4.0, 2.5)
Point2D(4.0, 2.5)
To change from segment coordinate to cartesian coordiantes, use the Point2D.where() method:
In [9]:
print(AB.where(I0))
print(CD.where(I0))
0.5
0.3333333333333333
Conlusion¶
This concludes the first tutorial part.
The Python interpreter has allowed us to interactively construct points, segments, and compute intersection between them. This a very handy way to perform temporary computation, using Python interpreter as an advanced calculator, or test a idea, write a prototype, etc.
The code can then be written to a Python script to be reused, and to build a library or an application. All the code will be interpreted and dynamically typed. If performance become an issue, we will want to add static typing. This is what will be covered in next section.
Using the Cython interface¶
geomalgo provides a cython API, making it easy to:
- compile code instead of interpret it,
- add static typing for extension types,
Cbuilt in, functions, etc,- compile to full performance
Ccode, usingC structuresinstead of extension types.
We first activate cython compilation in the notebook.
In [10]:
%load_ext Cython
%load_ext wurlitzer
The Cython extension is already loaded. To reload it, use:
%reload_ext Cython
Compiling the code¶
Instead of interpreting source code, cython compiles cython code to C code,
and the build a native Python module. Learn more about Cython.
For example, let’s rewrite the code in Cython. The %%cython magic do (learn more about
building cython code):
- compile Cython code to C code,
- compile C code to Python native module,
- execute the module (hence, the Cython code).
In [11]:
%%cython
import geomalgo as ga
A = ga.Point2D(2, 1, name='A')
B = ga.Point2D(6, 4, name='B')
C = ga.Point2D(5, 2, name='C')
D = ga.Point2D(2, 3.5, name='D')
AB = ga.Segment2D(A, B)
CD = ga.Segment2D(C, D)
I0, I1 = AB.intersect_segment(CD)
print(I0)
print(I1)
Point2D(4.0, 2.5)
None
Adding static typing¶
Once the code is written in Cython, which compile to C, static typing can be added.
The above code rewrittes to (notice the cimport instead of import):
In [12]:
%%cython
cimport geomalgo as ga
cdef:
ga.Point2D A, B, C, D
ga.Segment2D AB, CD
A = ga.Point2D(2, 1, name='A')
B = ga.Point2D(6, 4, name='B')
C = ga.Point2D(5, 2, name='C')
D = ga.Point2D(2, 3.5, name='D')
AB = ga.Segment2D(A, B)
CD = ga.Segment2D(C, D)
I0, I1 = AB.intersect_segment(CD)
print(I0)
print(I1)
Point2D(4.0, 2.5)
None
Using C structure instead of extension type¶
Calling extention type methods is as fast as calling C functions on C structure,
provided that extension type attributes and methods are all statically typed (cdef).
However, extension types can not be stack allocated as efficiently as C structure.
GeomAlgo relies on stack allocation for basic types (points, segments, etc).
For performance, we need be able to use C structures, instead of extension types.
At the end of this section, we will see that extension are wrapper around these C structures, and methods wrapper around C functions.
The above code is rewritten using CPoint2D, CSegment2D and
intersect_segment2d_segment2d().
In [13]:
%%cython
from libc.stdio cimport printf
cimport geomalgo as ga
cdef:
ga.CPoint2D A, B, C, D, I0, I1
ga.CSegment2D AB, CD
double coords[4]
int n
A.x, A.y = 2, 1
B.x, B.y = 6, 4
C.x, C.y = 5, 2
D.x, D.y = 2, 3.5
ga.segment2d_set(&AB, &A, &B)
ga.segment2d_set(&CD, &C, &D)
n = ga.intersect_segment2d_segment2d(&AB, &CD, &I0, &I1, coords)
printf("Number of intersection: %d\n", n)
printf("Intersection point: (%.2f, %.2f)\n", I0.x, I0.y)
Number of intersection: 1
Intersection point: (4.00, 2.50)
This generate “pure” C code, without reference to Python. It is as efficient as writting in C directly.
Relations between Python and Cython API¶
A Point2D is a wrapper around a CPoint2D, and coordinates are hold by CPoint2D.
![digraph Point2D {
Point2D -> "CPoint2D*"
"CPoint2D*" -> x
"CPoint2D*" -> y
"CPoint2D*" [fillcolor=gray,style="rounded,filled"]
x [fillcolor=gray,style="rounded,filled"]
y [fillcolor=gray,style="rounded,filled"]
}](_images/graphviz-668454c0f2d0daa6ea84b72a6a697bf1d8b7fda2.png)
For example:
In [14]:
%%cython
cimport geomalgo as ga
cdef:
ga.Point2D X
ga.CPoint2D* C_X
X = ga.Point2D(1, 2)
C_X = X.cpoint2d
- The Python API of a Segment2D extension type has two attributes:
- Point2D A (extension type)
- Point2D B (extension type)
- The Cython API has access to an additionnal attribute:
- CSegment2D (C structure)
- The CSegment2D struture has two members:
- CPoint2D* A (C structure)
- CPoint2D* B (C structure)
CSegment2D and Point2D A and B points to the same CPoint2D* A and B.
![digraph Segment2D {
Segment2D -> "CSegment2D"
Segment2D -> "Point2D A"
Segment2D -> "Point2D B"
CSegment2D -> "CPoint2D* A"
CSegment2D -> "CPoint2D* B"
"Point2D A" -> "CPoint2D* A"
"Point2D B" -> "CPoint2D* B"
"CPoint2D* A" -> "Ax"
"CPoint2D* A" -> "Ay"
"CPoint2D* B" -> "Bx"
"CPoint2D* B" -> "By"
"CSegment2D" [fillcolor=gray,style="rounded,filled"]
"CPoint2D* A" [fillcolor=gray,style="rounded,filled"]
"CPoint2D* B" [fillcolor=gray,style="rounded,filled"]
"Ax" [fillcolor=gray,style="rounded,filled"]
"Ay" [fillcolor=gray,style="rounded,filled"]
"Bx" [fillcolor=gray,style="rounded,filled"]
"By" [fillcolor=gray,style="rounded,filled"]
}](_images/graphviz-7094813e7642a0bdac9013b68db0ee42fc0e8002.png)
For example:
In [15]:
%%cython
cimport geomalgo as ga
cdef:
ga.Point2D X, Y
ga.Segment2D XY
X = ga.Point2D(1, 2)
Y = ga.Point2D(3, 4)
XY = ga.Segment2D(X, Y)
cdef:
ga.CSegment2D* C_XY
ga.CPoint2D* C_X
ga.CPoint2D* C_Y
C_XY = &XY.csegment2d
# Points can be accessed via X and Y
C_X = X.cpoint2d
# Or via C_XY
C_Y = C_XY.B
Working with large dataset¶
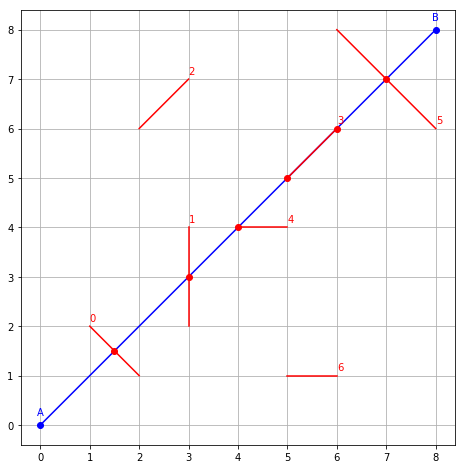
To conclude this tutorial, in this last part, the segment intersection computation will be applied to a (potentialy large) collection of segment.
In [16]:
A = ga.Point2D(0, 0, name='A')
B = ga.Point2D(8, 8, name='B')
AB = ga.Segment2D(A, B)
segment_list = [
ga.Segment2D((2,1), (1,2)),
ga.Segment2D((3,2), (3,4)),
ga.Segment2D((2,6), (3,7)),
ga.Segment2D((5,5), (6,6)),
ga.Segment2D((4,4), (5,4)),
ga.Segment2D((6,8), (8,6)),
ga.Segment2D((5,1), (6,1)),
]
figure(figsize=(8,8))
def plot_segments():
for obj in [A, B, AB]:
obj.plot()
for i, seg in enumerate(segment_list):
seg.plot(style='r-')
text(seg.B.x, seg.B.y+0.1, i, color='red')
plot_segments()
axis('scaled')
grid()
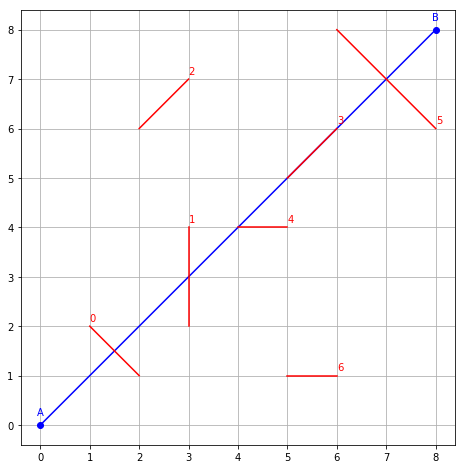
For example, we want to compute all intersection of red segments with the blue one, indexed by red segment indices. This should be optimized for a large number of red segments.
Memory layout¶
Segment coordinates will be stored as SoA. geomalgo provides a SegmentCollection extension type to groups the 4 arrays together.
In [17]:
xa = np.array([seg.A.x for seg in segment_list], dtype='d')
xb = np.array([seg.B.x for seg in segment_list], dtype='d')
ya = np.array([seg.A.y for seg in segment_list], dtype='d')
yb = np.array([seg.B.y for seg in segment_list], dtype='d')
segments = ga.Segment2DCollection(xa, xb, ya, yb)
A CSegment2D and its 2 CPoints2D are declared to iterate on the collection.
The Segment2DCollection.get is used to set PQ for the current segment index.
In [18]:
%%cython
from libc.stdio cimport printf
cimport geomalgo as ga
def iter_segments(ga.Segment2DCollection segments):
cdef:
ga.CSegment2D PQ
ga.CPoint2D P, Q
int S
ga.segment2d_set(&PQ, &P, &Q)
for S in range(segments.size):
segments.get(S, &PQ)
printf("(%.0f, %.0f), (%.0f, %.0f)\n", P.x, P.y, Q.x, Q.y)
In [19]:
iter_segments(segments)
(2, 1), (1, 2)
(3, 2), (3, 4)
(2, 6), (3, 7)
(5, 5), (6, 6)
(4, 4), (5, 4)
(6, 8), (8, 6)
(5, 1), (6, 1)
Computing intersection¶
Once we can iterate the segment collection, intersection can be computed as in the previous part with the intersect_segment2d_segment2d function.
We need to choose where to store the results. Each red segment may intersection the blue one on 0, 1 or 2 points. The total number of intersections points in not knonw in advance. We choose to allocate arrays to store results for the maximal number of intersection points. We also choose to store only the coordinates relative to the segment AB.
In [20]:
%%cython
import numpy as np
cimport geomalgo as ga
def compute_intersections(ga.Segment2D AB, ga.Segment2DCollection segments):
cdef:
ga.CSegment2D PQ
ga.CPoint2D P, Q
ga.CPoint2D I0, I1
double coords[4]
int S, C
# Count of intersections for each segment. May be 0, 1 or 2.
int[:] count = np.empty(segments.size, dtype='int32')
int countmax = 2
# Intersection coordinate relative to AB.
double[:,:] coord = np.empty((segments.size, countmax), dtype='d')
ga.segment2d_set(&PQ, &P, &Q)
for S in range(segments.size):
segments.get(S, &PQ)
count[S] = ga.intersect_segment2d_segment2d(&AB.csegment2d,
&PQ, &I0, &I1, coords)
for C in range(count[S]):
coord[S,C] = coords[C*2]
return np.asarray(count), np.asarray(coord)
Let’s call this function on our segments, and plot the result.
In [21]:
counts, coords = compute_intersections(AB, segments)
In [22]:
print(counts)
print(coords)
[1 1 0 2 1 1 0]
[[ 1.87500000e-001 2.64239983e-316]
[ 3.75000000e-001 0.00000000e+000]
[ 0.00000000e+000 0.00000000e+000]
[ 6.25000000e-001 7.50000000e-001]
[ 5.00000000e-001 0.00000000e+000]
[ 8.75000000e-001 0.00000000e+000]
[ 0.00000000e+000 0.00000000e+000]]
In [23]:
figure(figsize=(8,8))
plot_segments()
grid()
axis('scaled')
for S, (count, coord) in enumerate(zip(counts, coords)):
for C in range(count):
coord = coords[S, C]
P = AB.at(coord)
plot(P.x, P.y, 'ro')
Conlusion¶
In this tutorial, we’ve explored the Python and Cython API of geomalgo, and how to easily move from one to another depending on the needs.
Python API allows interactive processing using the Python scientific stack (numpy, matplotlib, jupyter notebook, etc), while Cython API give use C like performances.
Collection of object are stored as SoA, which can be iterate using a stack allocated element structure: for example, a Segment2DCollection is allocated using 4 arrays, and can be iterated using a CSegment2D structure to be processed.